Data Searches
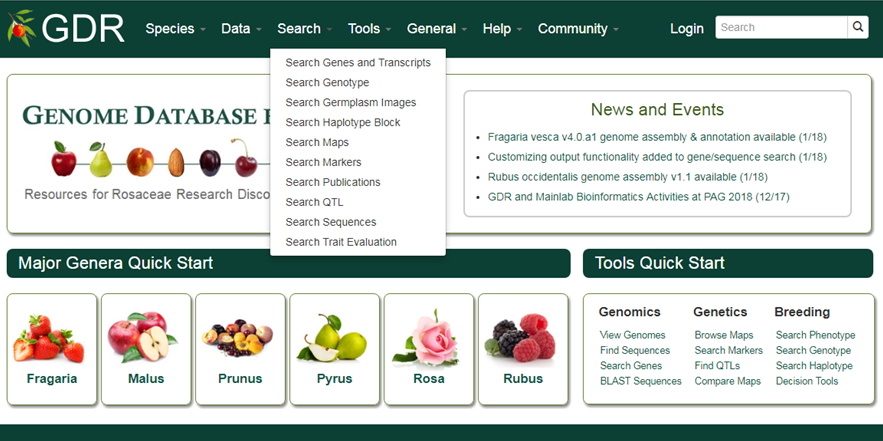
Search Genes and Transcripts
Search Genes and Transcripts is a page where users can search for genes and transcripts from various datasets available in GDR. Users can search for genes from various datasets: predicted genes from whole genome assemblies,a single non-redundant list of Rosaceae genes with gene symbols (GDR Gene Database), or gene and mRNA sequences parsed out from NCBI nucleotide database. These genes and mRNAs parsed out from NCBI sequences are aligned to the reference whole genome sequences when possible. When expert-contributed information is available, these gene names are associated directly with the predicted genes from whole genome assembly. Users can also search from transcripts from RefTrans sets, reference transcriptome sets built from all publicly available transcripts, or EST unigene contigs. For more detail, refer to 'Description of Gene and Transcript Dataset' page.
Please Note: All the search categories below, except the file upload, can be combined.
1. Genus/Species
Use these drop-down menus to limit results to genes from a specific genus or species. Once a genus is chosen, species for the genus are dynamically populated in the species dropdown. Major genera are listed on top of the Genus list under 'Common Selections' and the rest are listed as 'All options' in alphabetical order.
2. Dataset
Use this drop-down menu to limt the results to sequences from a specific dataset. Go to 'Description of Sequence Dataset in GDR' for more details. Multiple options can be selected by holding down the "Ctrl" key.
3. Genome Location
Users can limit their results of predicted genes by their genome location. When a genome assembly is chosen in the drop-down menu next to 'Dataset', the corresponding chromosome or scaffold names are dynamically displayed in the 'Genome Location' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
4. Gene/Transcript Name
Users can search genes and transcripts by name for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu. The search is case-insensitive. Example gene names are MADS1, LFY2, or ppa027130m. Gene or transcript names in a file, separated by a new line, can be uploaded to do a batch search.
3. Keyword
Users can limit their result by associated funcitonal terms. Predicted genes from whole genome assembly and transcripts have been annotated with some of the followings: homology to genes of closely related or plant model species, InterPro protein domains, GO terms, KEGG pathway and ortholog terms. Users can enter any protein name (eg. polygalacturonase), KEGG term/EC number (eg. resistance, EC:1.4.1.3), GO term (eg. cell cycle, ATP binding), or InterPro term (eg. zinc finger) in the text box to limit the results with the entries that are associated with the functional annotation terms.
If you have any questions/comments/feedback about this search page, please let us know via the contact form.
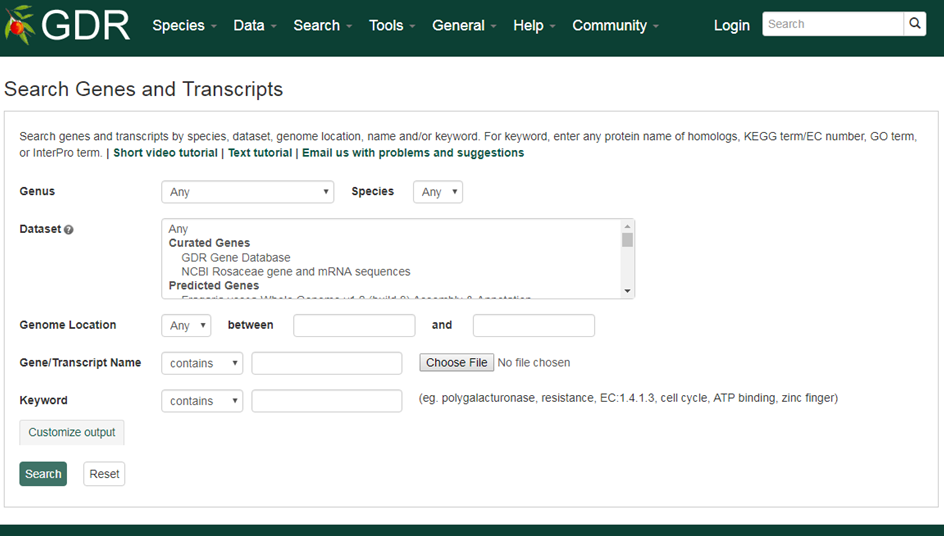
Search Genotype
Search SSR Genotype is a page where users can search the SSR genotype data by dataset name, marker name, germplasm name and/or species. Click the next tab to search for SNP Genotype. To search for SSR genotype data only for cultivars and breeding selections please visit the 'Search Genotyping Data' page in the Breeders Toolbox.
Please Note: All the search categories below can be combined.
1. Dataset
Users can search SSR genotype data by dataset from a dropdown list.
2. Marker Name
Users can search for SSR genotype data that used a specific marker by typing the marker name in the text box. Users can search for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu.
3. Germplasm Name
Users can search for SSR genotype data from a specific germplasm by choosing from the drop-down menu.
4. Species
Users can search for SSR genotype data from a specific species by choosing from the drop-down menu.
Search SNP Genotype is a page where users can search for the SNP genotyope dataset based on the germplasm and SNP markers used in the dataset. Click the next tab to search for SSR Genotype. To search for SNP genotype data only for cultivars and breeding selections please visit the 'Search Genotyping Data' page in the Breeders Toolbox.

Please Note: All the search categories below can be combined.
1. Dataset
Users can search SNP genotype data by dataset from a dropdown list.
2. Species
Users can search for SNP genotype dataset from a specific species by choosing from the drop-down menu.
3. Germplasm Name
Users can search for SNP genotype dataset from a specific germplasm by choosing from the drop-down menu.
4. SNP
Users can search for SNP genotype dataset that used a specific marker by typing the marker name in the text box. Users can search for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu.
5. Genome
Users can search SNP genotype data by the anchored position of SNPs in one of the whole genome sequences. Choose a genome in the drop-down menu next to 'Genome' then the corrposponding chromosome or scaffold names will by dynamically generated in the 'Chr/Scaffold' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
If you have any questions/comments/feedback about this page, please let us know via the contact form.
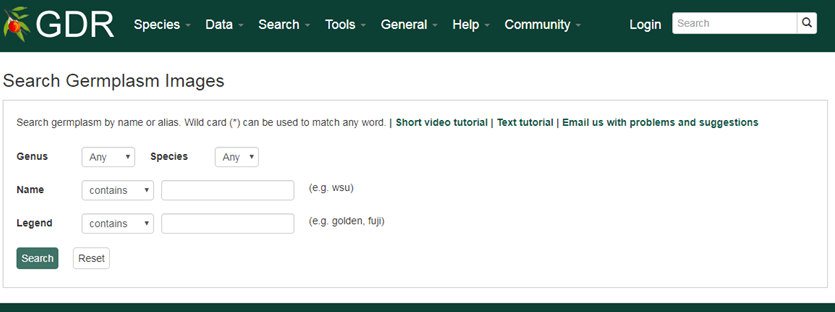
Search Germplasm Images
Search Germplasm Images is a page where users can search for germplasm images available in GDR. The search can be restricted by genus, species, germplasm name and the legend of the image.
Please Note: All the search categories below can be combined.
1. Genus/Species
Use these drop-down menus to limit results to genes from a specific genus or species. Once a genus is chosen, species for the genus are dynamically populated in the species dropdown. Major genera are listed on top of the Genus list under 'Common Selections' and the rest are listed as 'All options' in alphabetical order.
2. Name
Users can search images by germplasm name for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu. The search is case-insensitive.
3. Legend
Users can limit their result by associated legend of the image. Most images of Malus species have pedigree in their legends so users can search images by entering parent's name in the legend.
If you have any questions/comments/feedback about this search page, please let us know via the contact form.
Search Haplotype Block
Search Haplotype Block is a page where you can search for haplotype blocks, a genomic region which was identified to have a distinct combination of SNP genotype. The sesarch options include species, halotype block name or genome location to which the haplotype block is aligned. From the individual haplotype block page, you can view all the haplotype (alleles) identified along with the SNP genotypes that constitutes each genotype.
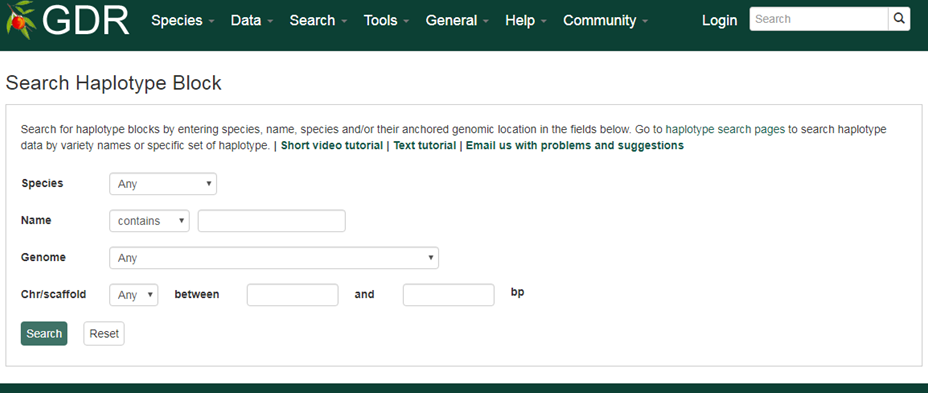
Please Note: All the search categories below can be combined.
1. Species
Use these drop-down menus to limit results to a specific species.
2. Name
Users can search by haplotype block name for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu. The search is case-insensitive.
3. Genome
Users can limit their results of haplotype blocks by their aligned genome location. When a genome assembly is chosen in the drop-down menu next to 'Genome', the corresponding chromosome or scaffold names are dynamically displayed in the 'Chr/scaffold' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
If you have any questions/comments/feedback about this search page, please let us know via the contact form.
Search Maps
Search Maps is a page where users can find and view Maps by species.
.png)
1. Species
Users can search for Maps from a specific species by choosing from the drop-down menu.
If you have any questions/comments/feedback about the marker search sites, please let us know via the contact form.
Search Publications
Search Publications is a page where users can search for publication using a combination of keywords (in the abstract or title), all or partial titles, authors, and other categories. Search results link to the publication detail pages that contain the abstract, citation, external link to the full article, and other details. The GDR houses information about publications on Rosaceae genomics, genetics, and breeding research. Details about publications were imported to the GDR from NCBI PubMed and the USDA National Agricultural Library using the query: (abstract: trait OR QTL OR gene OR genome OR map OR microsatellite OR annotation OR EST OR marker OR seqeuence) AND (abstract: rosaceae or prunus or pyrus or fragaria or malus or rubus). Additionally, details of publications from other journals not present in these databases are added.
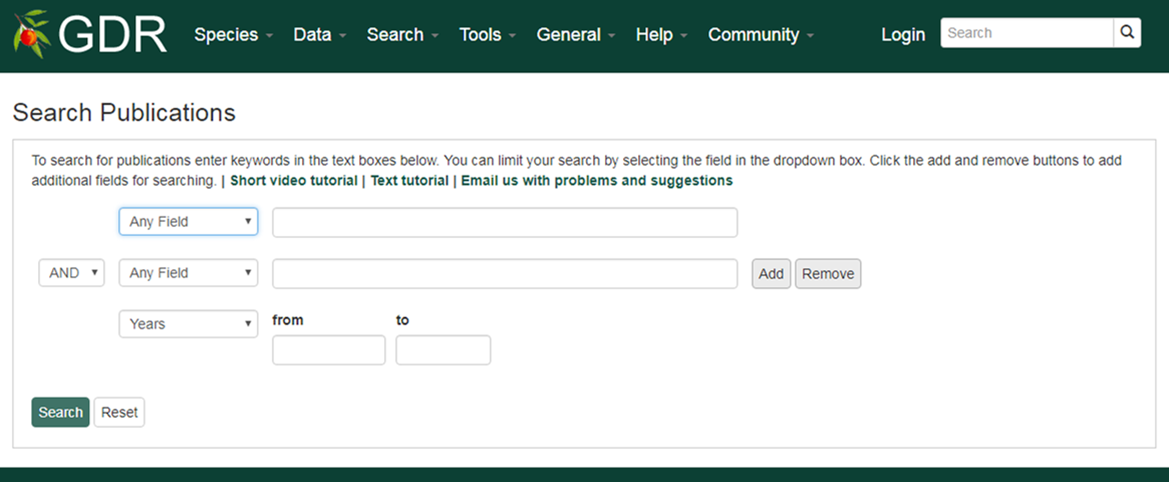
1. Users can select a field in the drop-down menu (abstract, authors, citation, journal name, title or year) and the type in keywords in the textbox
2. Users can expand their query by choosing 'AND, OR, NOT' and add additional field.
3. Users can click the plus symbol to add additional field.
4. Users can type in years in xxxx format limit the results between certain years.
If you have any questions/comments/feedback about the publications site, please let us know via the contact form.
Search QTL
Search QTL is a page where users can search for Quantatitive or Qualitative (Mendelian) Trait Loci in GDR.
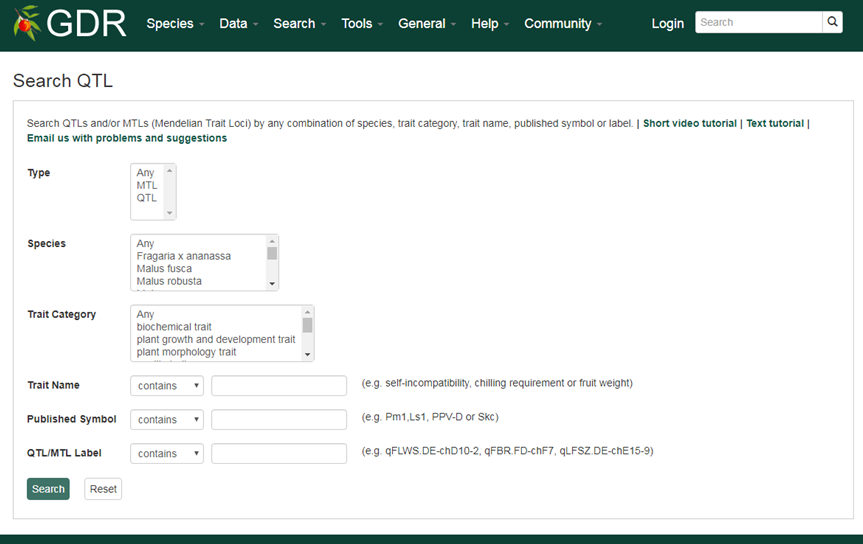
Please Note: All the search categories below can be combined.
1. Type
Users can search trait loci by type, QTL or MTL.
2. Species
Users can search trait loci by species by choosing one of the options displayed in the drop-down menu. Users can choose multiple options by holding down the "Ctrl" key.
3. Trait Category
Trait loci are associated with one or more terms of eight trait categories (anatomy and morphology, biochemical, growth and development, quality, stature or vigor, sterility or fetility, stress and yield). Users can choose multiple options by holding down the "Ctrl" key.
4. Trait Name
Trait loci are associated with trait names that belongs to the Rosaceae Trait Ontology. The Rosaceae Trait Ontology contains extra terms that are necessary for the description of Rosaceae traits in addition to the terms in Trait Ontology (developed with a focus on the traits of grass species). Examples include self-incompatibility, chilling requirement or fruit weight. Users can search these fields for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu. Users can also search by aliases. The search is case-insensitive.
5. Published Symbol
Published symbols can be used to search for trait loci. Examples include Pm1,Ls1, PPV-D or Skc. Users can search these fields for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu.
6. QTL/MTL Label
Search by the QTL/MTL label given by the GDR team.
An example QTL qSI.TE-ch5.1 indicates that:
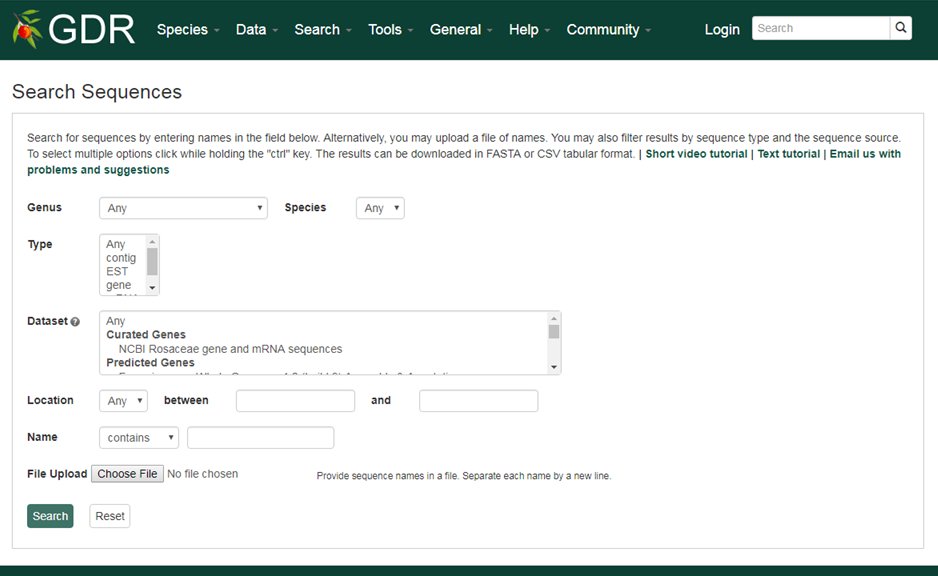
Search Sequences
If you have any questions/comments/feedback about the trait loci site, please let us know via the contact form.