Tools
To access the different Tools available on GDR, click on the 'tools' menu in the header and then select the tool you want to use. Many of the tools are also quickly accessed through links in the 'Tools Quick Start' section on the GDR homepage. To learn more about each search tool, please see the links below the figure.
How to view pan-genome data
How to view pan-genome data
To view available pangenome data click Pan-genome Data under Data dropdown.
Project page provide information on the pan-genome project as well as hyperlinks to the various pages and tools to acces the associated data. When pan-genome graph is available for the dataset, users can access the graph using the UCSC Genome Browser. Open the link for 'Pan-genome graph'.
Below is a snapshop of a UCSC Genome Browser showing pan-genome graph. Learn what the colors mean and some other useful hints.
JBrowse
GDR has an instance of the JBrowse genome browser for viewing genome data. A list of the genomes available in GDR can be accessed by clicking the JBrowse link in the Tools menu. Please watch the JBrowse tutorial for more details about how to navigate and use JBrowse.
Sequence Retrieval
The Sequence Retrieval tool allows downloading of nucleotide and protein sequences including chromosomes, scaffolds, genes, mRNAs, transcript coding sequences, protein, reftrans contigs and unigene contigs. For the sequences aligned to larger sequences, such as genes, mRNAs and transcript coding sequences, a numeric value specifying the number of upstream bases and downstream bases can be typed in the text boxes.
Below are currently available datasets for searching.
- Fragaria ananassa GDR RefTrans V1
- Fragaria Unigene v5.0
- Fragaria vesca Whole Genome v1.0 (build 8) Assembly & Annotation
- Fragaria vesca Whole Genome v1.1 Assembly & Annotation
- Fragaria vesca Whole Genome v4.0.a1 Assembly & Annotation
- Malus Unigene v5.0
- Malus x domestica GDDH13 v1.1 Whole Genome Assembly & Annotation
- Malus x domestica GDR RefTrans V1
- Malus x domestica Whole Genome v1.0 Assembly & Annotation
- Malus x domestica Whole Genome v1.0p Assembly & Annotation
- Prunus avium GDR RefTrans V1
- Prunus avium Whole Genome Assembly v1.0 & Annotation v1 (v1.0.a1)
- Prunus persica GDR RefTrans V1
- Prunus persica Whole Genome Assembly v2.0 & Annotation v2.1 (v2.0.a1)
- Prunus persica Whole Genome v1.0 Assembly & Annotation
- Prunus Unigene v5.0
- Pyrus communis Genome v1.0 Draft Assembly & Annotation
- Pyrus Unigene v5.0
- Rosaceae Family Unigene v5.0
- Rosa chinensis Whole Genome v1.0 Assembly & Annotation
- Rosa Unigene v5.0
- Rubus GDR RefTrans V1
- Rubus GDR RefTrans V2
- Rubus occidentalis Whole Genome Assembly v1.0 & Annotation v1
- Rubus occidentalis Whole Genome Assembly v1.1
- Rubus Unigene v5.0
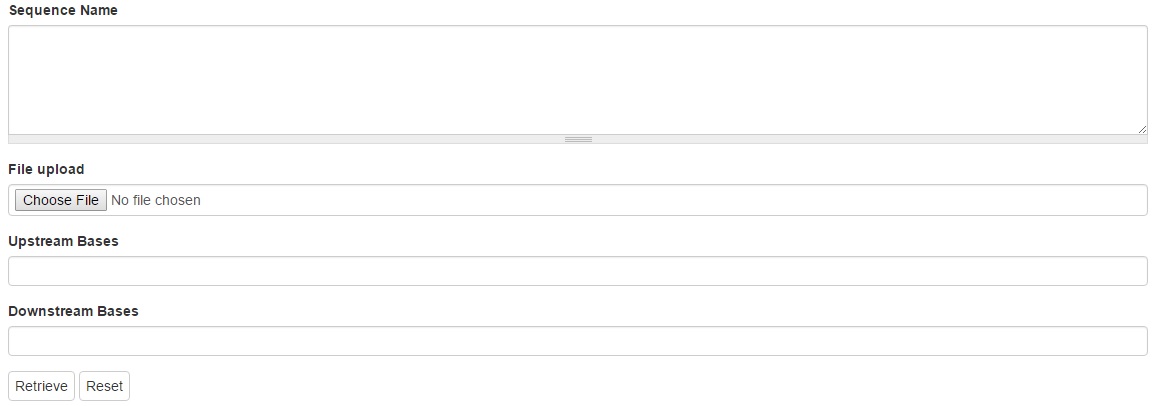
1. Sequence Name
The names of the sequences to be retrieved can be typed in the text box. Each name should be separated with a new line or comma. Leave blank to retrieve all features matching other criteria.
2. Upstream and downstream bases
A numeric value specifying the number of upstream bases and downstream bases to include in the downloaded sequences can be typed in the text boxes. This only works if the feature is aligned to a larger sequence.
If you have any questions/comments/feedback about the sequence retreival tool, please let us know via the contact form.
Synteny Viewer
Viewing RNA Expression Data
GDR has the Tripal Analysis Expression module to view differential expression values from RNA-seq experiments. Information about the different expression datasets on GDR is displayed in Expression Heatmap page. Expression data for the genome associated mRNAs or assembled transcriptome contigs is visualized either on the associated gene or mRNA page, or by creating a heatmap.
To view if your gene or mRNA of interest have associated expression data, search and find them in "Search Gene and Transcripts". Genes of mRNAs that have expression data will have an "Expression" category on the left hand menu of the feature page.
IMAGE
When "Expression" is clicked, the center pane will change to display the available expression data. Hovering over the bars on the chart displays the expression value and more infomation about the sample. Clicking on the bars will open the information page for the biomaterial, which has links to the analysis details page. There are also options to sort or edit the display of the expression data. When the gene/mRNA is associated with more than one analysis page, a dropdown will show up so that users can change the analysis.