Search Sequences
The Search Sequences page allows users to search for various sequences in the GDR database. These sequences include genes and transcripts from various sources, RosCOS unigene sets, RosaR80 v1.0. Please follow the link for more information on each source.
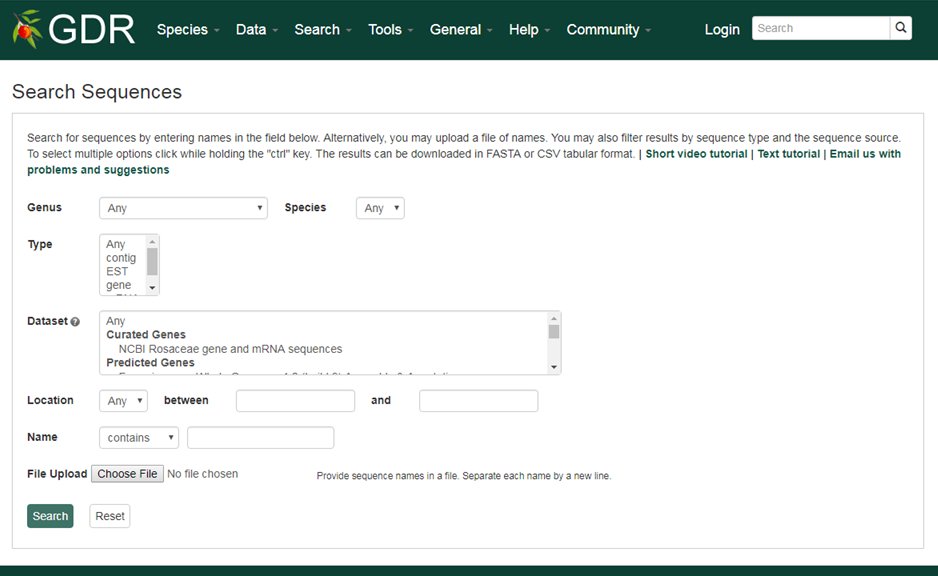
Please Note: All the search categories below, except the file upload, can be combined.
1. Species
Use this drop-down menu to limit results to sequences from a specific species. Multiple options can be selected by holding down the "Ctrl" key.
2. Type
Use this drop-down menu to limit results to sequences of a specific type. Multiple options can be selected by holding down the "Ctrl" key.
3. Dataset
Use this drop-down menu to limt the results to sequences from a specific source: genes and transcripts from various sources, RosCOS unigene sets, RosaR80 v1.0. Please follow the link for more information on each source. Multiple options can be selected by holding down the "Ctrl" key.
3. Name
Users can search sequences by name for an exact match, contains, starts with or ends with the input, by selecting the desired option from the drop-down menu. The search is case-insensitive.
4. File Upload
Users can obtain detailed information for a set of sequences by uploading a file with sequences names. Separate each name by a new line.
5. Location
Users can limit their results to those aligned to a specific genome assembly. In addition to the predicted genes and mRNAs from each assembly, NCBI Rosaceae genes and mRNA sequences are aligned to the reference whole genomes with criteria of >98% PID and >95% Aligned Length. When a genome assembly or 'NCBI Rosaceae genes and mRNA sequences' is selected in the drop-down menu next to 'Dataset', the corrposponding chromosome or scaffold names will be dynamically displayed in the 'Location' drop-down menu. Choose any option and then type in the position in bp in the text boxes.
If you have any questions/comments/feedback about the trait loci site, please let us know via the contact form.